Corona Virus
Corona
8 June 2020
there is better understanding about Corona now, Ventilators did not kill, Doctors thought Corona infected always die of Lung failure, so ventilator is best aid ,they left aside other things in absense of any info n data...so many patients died... Post mortem info started flowing from Italy n USA, they found most of people died of heart attack... N it's most normal in any kind of severe infection ,doctors realized it ,change the course of treatment... Most of elderly people have gun ready in their body n it needs just trigger...that trigger is high toxicity due to infection...hyper tension, diabetes both are connected with heart problem.... Hyper tension indicate that gun is ready n diabetes make sure reduced kidney function allowing toxicity to build in body... Another very important factor was known at later stage that reduced level of D vitamin triggers Cytokine storm when infected by Corona, Cytokine storm means over reaction of body immune system which start killing body itself, Effect of that is renal failure n heart failure.... So people with high level of morbidity should be careful... Didn't I tell you to take D vitamin with C vitamin ??Virus
| Virus | A virus is a microscopic package of genetic material surrounded by a molecular envelope. The genetic material can be either DNA or RNA. |
| DNA | DNA is a two-strand molecule that is found in all organisms, such as animals, plants, and viruses, and it holds the genetic code, or blueprint, for how these organisms are made and develop. |
| RNA | RNA is generally a one-strand molecule that copies, transcribes and transmits parts of the genetic code to proteins so they can synthetize and carry out functions that keep organisms alive and developing. There are different variations of RNA that do the copying, transcribing and transmitting. |
| Corona | Some viruses such as the coronavirus (SARS-Cov2) only contain RNA, which means they rely on infiltrating healthy cells to multiply and survive. Once inside the cell, the virus uses its own genetic code RNA in the case of the coronavirus to take control of and ‘reprogramme’ the cells so that they become virus-making factories. |
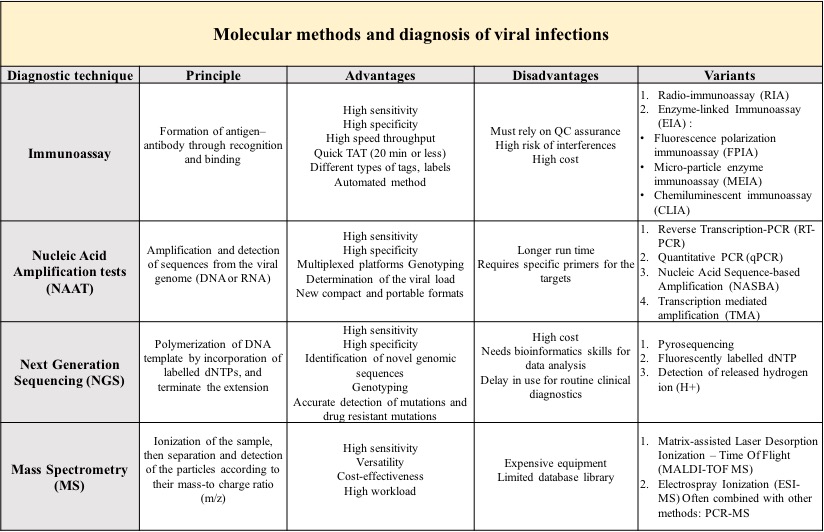
| Dignosis | Laboratory diagnosis of viral infections comprises a spectre of methods and techniques with different sensitivity and specificity that are used in routine and research laboratory work. Modern laboratory procedures in virology can be divided in two groups: conventional and molecular methods. |
Conventional
Conventional laboratory methods for diagnosis of viral infections -
- For long time, clinical laboratories have relied on a diverse range of techniques to diagnose viral infections.
- In developed countries, electron microscope (EM) and Immunofluorescence (IF) has for long time been considered an efficient tool for direct detection of viruses through visualization and counting of the viral particles in body fluids, stools or histopathologic samples.
- The identification is based on morphological characteristics specific to each virus family and requires a certain amount of viral particles (up to 10^6 particles/ml).
- However, specimen preparation that must be performed beforehand may reduce the virus concentration which makes the analysis harder.
- The time required for isolating viruses by cell culture is very long (weeks), limiting the usefulness of this technique when rapid diagnosis is needed.
The above-mentioned conventional methods have lower sensitivity and unreliability as disadvantages when compared to modern, molecular methods.
Molecular - Advantages
- High sensitivity and specificity assays approaching 100%.
- Possibility to combine different methods in one assay enhances detection capability and accuracy (PCR-MS, microfluidic chip technology-based PCR,).
- Automation of the assays reduces the number of operators and manual workload.
- Small sample volumes are needed, so the assays can still be performed in particular cases (CSF, new-borns, etc.)
- Some assays such as MS remain efficient after several freezingthawing of the samples (archived samples).
- Rapid Turn Around Time: minimum time required from sample collection to results reporting is 30 min, and microarray tools can deliver results within seconds.
- Low detection limit: 10100 copies/ml by PCR.
- Multiplex reactions: detection of a wide range of pathogens in a single run is time saving.
- Detection of rare drug-resistance patterns.
- NGS generates many sequencing data per run and sequences long-reads.
- Low reaction cost when using microfluidic chips, and use in Point-Of-Care settings.
- Lower risk of contamination by processing single tube within one unit.
RT-PCR - Why?
- In order for a virus like the coronavirus to be detected early in the body using real time RT-PCR, scientists need to convert the RNA to DNA.
- This is a process called ‘reverse transcription’.
- They do this because only DNA can be copied or amplified which is a key part of the real time RT-PCR process for detecting viruses.
- Scientists amplify a specific part of the transcribed viral DNA hundreds of thousands of times.
- Amplification is important so that instead of trying to spot a minuscule amount of the virus among millions of strands of genetic information, scientists have a large enough quantity of the target sections of viral DNA to accurately confirm that the virus is present.
- The real time RT-PCR technique is highly sensitive and specific and can deliver a reliable diagnosis as fast as three hours, though usually laboratories take on average between 6 to 8 hours.
- Compared to other available virus isolation methods, real time RT-PCR is significantly faster and has a lower potential for contamination or errors as the entire process can be done within a closed tube.
- It continues to be the most accurate method available for detection of the coronavirus.
RT-PCR - How?
- A sample is collected from parts of the body where the coronavirus gathers, such as a person’s nose or throat.
- The sample is treated with several chemical solutions that remove substances, such as proteins and fats, and extracts only the RNA present in the sample.
- This extracted RNA is a mix of a person’s own genetic material and, if present, the coronavirus’ RNA.
- The RNA is reverse transcribed to DNA using a specific enzyme.
- Scientists then add additional short fragments of DNA that are complementary to specific parts of the transcribed viral DNA.
- These fragments attach themselves to target sections of the viral DNA if the virus is present in a sample.
- Some of the added genetic fragments are for building DNA strands during amplification, while the others are for building the DNA and adding marker labels to the strands, which are then used to detect the virus.
- The mixture is then placed in a RT-PCR machine.
- The machine cycles through temperatures that heat and cool the mixture to trigger specific chemical reactions that create new, identical copies of the target sections of viral DNA.
- The cycle repeats over and over to continue copying the target sections of viral DNA. Each cycle doubles the previous amount: two copies become four, four copies become eight, and so on.
- A standard real time RT-PCR setup usually goes through 35 cycles, which means that by the end of the process, around 35 billion new copies of the sections of viral DNA are created from each strand of the virus present in the sample.
- As new copies of the viral DNA sections are built, the marker labels attach to the DNA strands and then release a fluorescent dye, which is measured by the machine’s computer and presented in real time on the screen.
- The computer tracks the amount of fluorescence in the sample after each cycle.
- When the amount goes over a certain level of fluorescence, this confirms that the virus is present.
- Scientists also monitor how many cycles it takes to reach this level in order to estimate the severity of the infection: the fewer the cycles, the more severe the viral infection is.
Antigen/Antibody
- In case of an infection, the antigens present on the virus elicits an immune response in the body.
- This response triggers a cascade mechanism for the development of antibodies against the antigens.
- The produced antibodies are isolated from the human blood or plasma and are introduced into a mice.
- The antibodies from a human system are detected as foreign entities by the mice immune system and they start producing antibodies against the antibodies from the human system.
- These newly produced antibodies are called as anti-antibodies or monoclonal antibodies.
- Anti- antibodies from the mice bind to a secondary antibody that are labelled with a marker for identification purposes.
- This anti-antibody and tagged secondary antibody complex is coated on the test paper strip.
- On addition of the sample for detection of infection(test sample), antibodies from the test sample will bind to the anti- antibodies and due to the marker on the secondary antibody, a response will be exhnibited. Thus, giving out results in visual form.
Last Edit: Mon Mar 24 19:59:18 2025, EDT